Article Text
Abstract
Objective Invasive meningococcal disease (IMD) outbreaks in men who have sex with men (MSM) have been associated with meningococcal colonisation of the urethra and rectum, but little is known about this colonisation or co-colonisation with the closely related gonococcus. Whole genome sequencing (WGS) was employed to explore these phenomena.
Methods Meningococci isolated from the urogenital tract and rectum (n=23) and coincident gonococci (n=14) were analysed by WGS along with contemporary meningococci from IMD (n=11). All isolates were obtained from hospital admissions in Brighton, UK, 2011–2013. Assembled WGS were deposited in the PubMLST/neisseria database (http://pubmlst.org/neisseria) and compared at genomic loci common to gonococci or meningococci.
Results As expected, most meningococci from IMD were encapsulated and belonged to hyperinvasive lineages. So too were meningococci found in the urogenital tract and rectum, contrasting to those asymptomatically carried in the nasopharynx where such meningococci are rare. Five hyperinvasive meningococcal lineages and four distinct gonococcal genotypes were recovered, including multiresistant ST-1901 (NG MAST-1407) gonococci.
Conclusions These data were consistent with a predisposition for potentially virulent encapsulated hyperinvasive meningococci to colonise the urethra and rectum, which suggests their involvement in MSM IMD outbreaks. The coincidence of multiresistant gonococci raises wider public health concerns.
- NEISSERIA GONORRHOEA
- NEISSERIA MENINGITIS
- INFECTIOUS DISEASES
- MENINGITIS
This is an Open Access article distributed in accordance with the terms of the Creative Commons Attribution (CC BY 4.0) license, which permits others to distribute, remix, adapt and build upon this work, for commercial use, provided the original work is properly cited. See: http://creativecommons.org/licenses/by/4.0/
Statistics from Altmetric.com
Introduction
Neisseria meningitidis and Neisseria gonorrhoeae are two closely related Gram-negative diplococcal bacteria responsible for invasive meningococcal disease (IMD) and gonorrhoea, respectively. Their genetic similarity indicates they diverged from the same ancestral population in the relatively recent past.1 Given the much lower diversity of the gonococcus, it is thought to have evolved by a change of niche from the nasopharynx, where most Neisseria are found, to the urogenital tract, but paradoxically, reports of clinically recognisable gonococcal disease are much older (1376, and perhaps much earlier)2 than meningococcal disease, which is widely considered to have been first recognised in 1805.3
Meningococcal disease can be thought of as an emerging infectious disease of the early 1800s, occurring in a growing human population, which, at the beginning of the 19th century, had exceeded 1 billion.4 IMD is predominantly caused by meningococci expressing a polysaccharide capsule, the biochemical and genetic composition of which determine the serogroup, with six serogroups (A, B, C, W, X and Y) causing almost all IMD.5 In addition, most invasive meningococci belong to fewer than 20 genotypes, known as ‘hyperinvasive lineages’, which are identified, by multilocus sequence typing (MLST), as groups of related sequence types (STs) called clonal complexes (ccs).6 ,7 Almost all infections caused by meningococci result from asymptomatic carriage and IMD is inimical to spread as it does not result in transmission.
While the human nasopharynx remains the site that is preferentially colonised by N. meningitidis, an increasing number of urogenital and rectal meningococcal infections have been reported, accompanied by evidence of transmission between sexual partners involving the nasopharynx, urogenital tract and rectum.8 ,9 These transmission pathways may have facilitated the IMD outbreaks recently reported among men who have sex with men (MSM) in Canada, the United States, Germany, France and Belgium.10–13 These outbreaks were associated with serogroup C meningococci belonging to the hyperinvasive ST-11 clonal complex, prompting the use of vaccination with the quadrivalent ACWY conjugate vaccine as a public health response.14 Although these outbreaks indicate that urogenital and rectal colonisation by N. meningitidis may increase opportunities for transmission and infection, little is known about the prevalence and strain characteristics of meningococci in the urogenital tract and rectum. Here, we present genomic analyses and comparison of such organisms from Brighton, UK, which were temporally matched with whole genome sequencing (WGS) data obtained from cases of IMD from the same city. The increasing presence of meningococci in the urogenital tract may represent a niche into which this organism is expanding, perhaps leading to the emergence of novel infectious variants that mirror the emergence of gonococcal disease and this warrants surveillance.
Materials and methods
Clinical setting and samples
Isolates originated from routine diagnostic samples submitted to microbiology at the Brighton and Sussex University Hospitals NHS Trust (BSUH) Brighton, UK, calendar years 2011–2013. N. meningitidis isolates were obtained from acute medical patients with invasive disease and also from patients attending sexual health services (c25000 attendances/year, 25% MSM) in whom N. meningitidis was isolated incidentally during routine urethral and rectal screening for gonorrhoea. During the study period, 11 meningococci were obtained and archived from cases of invasive disease (7 blood culture, 2 synovial fluid, 2 cerebrospinal fluid): information on the sexual orientation was not available. During the same period, 36 urogenital/rectal meningococci were detected, of which 7/36 were urethral and 29/36 were rectal samples: 23/36 viable urogenital/rectal N. meningitidis isolates were available for further study. As pharyngeal meningococci were not considered clinically significant, they were not routinely archived. Between 2011 and 2013, a total of 1033 N. gonorrhoeae isolates were recorded with 12 patients (all male) yielding both N. meningitidis and N. gonorrhoeae (5 concurrently, 7 more than a month apart). Fourteen gonococci from these patients were available for study.
N. gonorrhoeae isolates were initially identified using a nucleic acid amplification test (BD ProbeTec, BD, Franklin Lakes, New Jersey, USA).15 N. gonorrhoeae genomic DNA was extracted from isolates subcultured onto VCAT (Vancomycin, Colistin sulfate, Amphotericin B, Trimethoprim) selective agar (Oxoid, Basingstoke, UK) and incubated in a CO2-rich atmosphere at 37°C for 24 hours. Prior to DNA extraction, N. meningitidis isolates were streak plated onto Columbia agar plus 5% (v/v) sheep blood and incubated overnight at 37°C in an atmosphere containing 5% CO2. Gonococci and meningococci were stored in 20% glycerol broth at −80°C. Antimicrobial susceptibilities were determined using agar diffusion with Etest and published British Society for Antimicrobial Chemotherapy (BSAC) guidelines for minimum inhibitory concentration (MIC) interpretation.16
WGS and analysis
Genomic gonococcal DNA was extracted using a commercial kit (QuickGene, Fujifilm, Tokyo, Japan). Meningococcal genomic DNA was extracted using the Wizard Genomic DNA purification kit (Promega). Both meningococci and gonococci were sequenced using the Illumina HiSeq platform and short-read sequences were assembled de novo using the VelvetOptimiser assembly program. Resultant assemblies were uploaded to the pubMLST database running the BIGSdb genomic platform hosted on http://www.pubMLST.org/neisseria. WGS data from a French cc11 serogroup C meningococcus (LNP27256), originating from a French MSM outbreak, were also included in the analyses as well as the genome from a cc4821 N. meningitidis Chinese isolate, 053442.12 ,17
PubMLST.org/neisseria archives and annotates, at the time of writing, >7000 WGS data from across the Neisseria genus. WGS deposited in the database are automatically annotated for any defined loci, identifying alleles ≥98% sequence identity and updating isolate records with allele numbers. This enabled the genogroup, PorA and FetA type as well as MLST ST or NG MAST to be identified. Chromosomal and plasmid genes in addition to intergenic regions implicated in gonococcal antimicrobial resistance (AMR) have also been defined in the pubMLST Neisseria database with alleles containing known mutations associated with AMR annotated accordingly.18 ,19
The BIGsdb Genome comparator tool, implemented within the website, was employed to compare WGS data.20 Using this tool, 1605 loci identified as core to meningococci and belonging to the N. meningitidis core genome MLST (cgMLST) scheme V.1.0 were compared between all meningococci.21 In addition, 1668 loci, identified as core to gonococci in this dataset and defined in the N. gonorrhoeae cgMLST scheme V.1.0, were compared between all gonococci. Using the genome comparator tool, coding sequences from selected loci are extracted and compared against assembled WGS data. Alleles at each locus are designated with an integer, identifying isolates with the same or different allelic profiles, and a distance matrix is generated based on the number of variable alleles resolving isolates into networks using the NEIGHBORNET algorithm and a standalone instance of SPLITSTREE.22 ,23 Gene-by-gene comparisons undertaken using genome comparator also provide lists of loci identical, variable, missing or incomplete between datasets thereby resolving population relationships and elucidating where genetic variation is taking place. This in turn allows the functional significance of such variation to be identified.
Results
Isolate characterisation
During the study period, N. meningitidis was obtained more frequently from rectal (19/23, 83%) than urethral samples (4/23, 17%) but was detected less frequently than N. gonorrhoeae (365/1033, 32% rectal; 435/1033, 47% urethral; 185/1033, 16% pharyngeal; 45/1033, 4% endocervix; 3/1033, 0.2% other sites including conjunctiva and intrauterine contraceptive device). Thirteen (56%) of the 23 urogenital N. meningitidis isolates belonged to clonal complexes associated with IMD (table 1). There were also clonal complexes more commonly found in carriage, for example, cc1157 (n=3), and isolates with STs not currently associated with a clonal complex (table 1). Isolates from clonal complexes not associated with IMD were either unencapsulated, possessing the capsule null locus (cnl) or were serogroups E or Z. Most of the remaining isolates were serogroup B (67%) belonging to clonal complexes cc41/44, cc269, cc4821 as well as the ST-1976 isolates, while the cc11 isolates were serogroup C and the cc23 isolates were serogroup Y. There were three cc1157 isolates and these were serogroup B, E or Z (table 1).
Isolate collection
The 14 N. gonorrhoeae isolates corresponded to 2 ST-9363 (NG-MAST ST-2992) isolates (one urethral, one rectal) obtained from one patient on the same day; 5 ST-7363 (NG-MAST ST-2400) or ST-11516 (new NG-MAST) isolates obtained from five individuals between 2011 and 2013; 3 ST-1901 (NG-MAST ST-1407) isolates, 2 of which (pharyngeal and rectal samples) were obtained on the same day from the same patient and a pharyngeal isolate from another patient.
Antimicrobial resistance
None of the meningococci exhibited phenotypic resistance to fluoroquinolones or macrolides according to the BSAC guidelines (see online supplementary table S1);16 however, 15 isolates contained NEIS1753 (penA) alleles with known amino acid substitutions associated with resistance to penicillin.22 Of these, 14/15 showed reduced susceptibility phenotypically to penicillin (MIC range 0.13–0.5). Three isolates displayed reduced susceptibility to penicillin phenotypically but did not contain a corresponding resistant genotype (MIC range 0.25–0.5).
supplementary data
Four of the 14 N. gonorrhoeae isolates (29%) were resistant to β-lactams, including third-generation cephalosporins and fluoroquinolones (data not shown). Three isolates without mosaic penA alleles exhibited a deletion in the promoter region of the efflux pump regulator, mtrR, associated with increased expression of the efflux pump. In the remaining seven isolates, genotypic resistance markers were not detected. Neither mutations in 16S rRNA or 23S rRNA conferring resistance to spectinomycin or azithromycin, respectively, nor plasmid-mediated AMR was detected in any isolate.
N. meningitidis genomic analyses
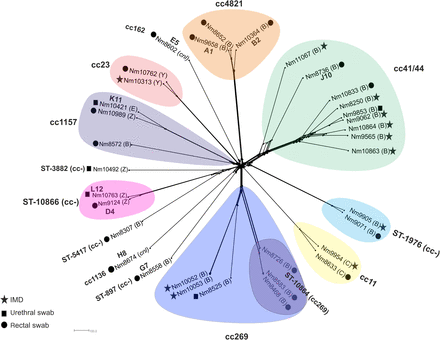
Genomic comparison of meningococcal core loci were phylogenetically reconstructed with isolates clustering by lineage. Of the eight clusters, five included both invasive and urogenital/rectal isolates (figure 1). Locus differences were used to assess relatedness and possible involvement in transmission networks, with differences in ≤20 loci being considered compatible with recent transmission.24 Two ST-1976 isolates (figure 1), NM9071 (rectum) and NM9905 (IMD), showed matching strain designation (B: P1.22-1,14: F5-2) with cgMLST analysis identifying 130 locus differences.
Neisseria meningitidis genome comparison. NeighborNet tree depicting N. meningitidis isolates compared using 1605 loci core to the meningococcal genome (N. meningitidis cgMLST V.1.0). Stars depict invasive meningococcal disease cases; black squares N. meningitidis isolates obtained from urethral samples; black circles N. meningitidis isolates retrieved from rectal swabs. A1, B2, D4 and so on indicate concomitant N. gonorrhoeae infections. (B) serogroup B, (C) serogroup C, (E) serogroup E, (Y) serogroup Y, (Z) serogroup Z, (cnl) capsule null. IMD, invasive meningococcal disease
The clonal complex cc41/44 isolates, NM9062 (blood culture) and NM9853 (urethral), (figure 1) exhibited the same strain designation (B: P1.7-2,4: F1-5: ST-41) and contained distinctive alleles not found in the other cc41/44 isolates in this collection. NM8736 was distinct from other cc41/44 strains in this study (see online supplementary figure S1A).
CC269 isolates, NM8468, NM8583 and NM8726, all with the same strain designation (B: P1.19,15-1: F1-5: ST-10864 cc269) and all rectal isolates, formed a distinct cluster (figure 1). NM8468 and NM8583 differed in 4 loci, whereas NM8726 differed from these in a further 108 loci. NM10052 and NM10053 also from cc269, but from IMD, differed in 12 loci. These isolates were distinct from the other cc269 strains. Divergence was apparent in isolates NM9124 (rectum) and NM10763 (urethra) although both of these had the same strain designation (Z:P1.22, 14-23:F5-7:ST-10866) (figure 1). These isolates differed in 122 loci, including loci associated with energy and DNA metabolism, hypothetical proteins and loci implicated in pilin biogenesis.
The French MSM outbreak meningococcus, LNP27256, had the same strain designation as isolate NM8633 (C:P1.5-1,10-8:F3-6:ST-11)12 but exhibited 114 locus differences, whereas 313 locus differences were apparent between these and NM9954 (IMD non-MSM). The gene, aniA, encoding nitrate reductase and facilitating persistence in anaerobic environments, was found to be functional in all but 8/36 (22%) isolates in this dataset (table 1).25
Isolates NM103364, NM8652 and NM9658 belonged to cc4821, which is associated with IMD in China and one case of cc4821 IMD (isolate M14-240580) has been identified in the UK in 2014 (MRF Meningococcus Genome Library http://www.meningitis.org/research/genome). These isolates were distinct from the Chinese isolate, 054332 (see online supplementary figure S1B) and isolates NM10364 and M14-240580 clustered separately from other cc4821 meningococci, consistent with their different PorA types. A total of 362 locus differences were observed between the two clusters with 108 locus differences found between NM10364 and M14-240580.
N. gonorrhoeae genome analyses
Core genome analysis identified four distinct clusters (figure 2). These included the ST-1901 genotype, comprising four isolates, one of which ST-7360; the ST-7363 and ST-9363 lineages containing three isolates each and the ST-11516 lineage including two isolates. The remaining two isolates were located on long branches distantly related to ST-11516.
Neisseria gonorrhoeae genome comparison. NeighborNet tree depicting N. gonorrhoeae isolates compared using the 1668 loci core to the gonococcal genome (N. gonorrhoeae cgMLST V.1.0). Black squares depict isolates obtained from urethral samples; black circles isolates retrieved from rectal swabs with triangles indicating nasopharyngeal isolates. Red circles depict gonococci resistant to multiple antimicrobial compounds including third-generation cephalosporins and fluoroquinolones; pink circles indicate gonococci resistant to fluoroquinolones and penicillin, while green circles depict antimicrobial susceptible gonococci.
A total of 106 locus differences were observed between isolates in the ST-1901 lineage. This decreased to 25 locus differences after exclusion of the ST-7360 isolate, decreasing to 2 locus differences (15 polymorphic sites due to an indel in a hypothetical protein) once the 2 ST-1901 isolates from patient J10 were compared alone. Patient J10 also had a urethral N. gonorrhoeae infection; however, this was due to a genomically distinct ST-11516 isolate (figure 2). A total of 72 locus differences were observed between isolates in the ST-7363 cluster. This decreased to 22 locus differences when isolates from patients E5 and G7 only were compared. One variant locus, due to a frame-shift in a homopolymeric tract of the gene hpuA implicated in iron acquisition from haemoglobin–haptoglobin, was observed between gonococci from patient A1 (rectum and urethra).
Discussion
Urogenital and rectal meningococcal infections have been increasingly reported since the 1930s,26 with recent accounts associating meningococci expressing serogroup C capsules with IMD outbreaks among MSM.8 ,9 In this study, the majority of urogenital and rectal meningococci analysed belonged to hyperinvasive lineages (table 1, figure 1). This contrasts with the situation in nasopharyngeal carriage, where the prevalence of hyperinvasive lineages is lower.27 Meningococcal carriage is dominated by less invasive meningococci, which are frequently acapsulate or express capsules not associated with IMD. This suggests that common genetic determinants may be involved in promoting both urethral/rectal colonisation and IMD. As the rectum and urethra are physiologically distinct from the nasopharynx, factors such as the polysaccharide capsule, a well-known meningococcal virulence determinant, may be important for colonisation and persistence (table 1), consistent with the prevalence of encapsulated strains identified here and previously.28–30
The expansion of meningococci into this environment may result in adaptation leading to the emergence of strains that are able to persist by novel means of colonisation and transmission. An example is the gene aniA encoding nitrite reductase and essential for the growth of gonococci under oxygen-limiting conditions. Both urogenital and invasive cc11 N. meningitidis isolates from MSM have been found to express this gene as opposed to invasive strains obtained from non-MSM cases, which do not.25 Gene-by-gene comparisons undertaken here between MSM and non-MSM derived cc11 meningococci identified further variation in genes implicated in the respiratory chain as well as Na+ translocating NADH-ubiquinone reductase subunits, which are essential for ATP synthesis. These changes are consistent with adapative metabolic changes that may promote adaptation. Further comparisons between pairs of invasive and urogenital/rectal meningococci from the same lineage, identified an average of 125 locus differences, comprising genes associated with core metabolic functions such as amino acid biosynthesis, DNA metabolism and central intermediary metabolism. Such changes may promote adaptation to the urogenital tract.
Until now, there has been relatively little interest in the population dynamics of urogenital/rectal meningococci. Our findings, however, indicate that investigation of such strains is warranted, particularly as urogenital and rectal carriage may promote the introduction and transmission of potentially invasive meningococci. For example, both of the ST-1976 strains described shared the same PorA, FetA genes with only nine other isolates with this ST recorded in the PubMLST database at the time of writing, none of which from the UK. We also identified three urogenital/rectal serogroup B cc4821 meningococci (table 1). IMD outbreaks due to cc4821 meningococci were previously limited to Asia, particularly China; however, a case of IMD due to cc4821 was identified for the first time in the UK in 2014 {MRF Meningococcus Genome Library http://www.meningitis.org/research/genomehttp://www.meningitis.org/research/genome, #7406}. The strain designation of this isolate was identical to the cc4821 rectal strains, but distinct from Chinese cc4821 meningococci (see online supplementary figure S1B).31 The continued presence of cc4821 in the UK indicates that it should be monitored, since coverage with the serogroup B vaccines, Bexsero and Trumenba, has not been determined for strains from this clonal complex.32
As potential pathogens, urogenital and rectal meningococci pose a risk to public health. Their frequent coexistence with gonococci could promote the transfer of AMR genes (figure 2). This is particularly important in gonococci exhibiting decreased susceptibility to third-generation cephalosporins mediated through recombination of mosaic penA alleles. The annotation of genes across the Neisseria genus enabled by pubMLST.org/neisseria uniquely allows identification of horizontal genetic transfer events between species. We found evidence of such events; for example, N. meningitidis isolate NM8633 possessed a penA allele more commonly found in gonococci (see online supplementary table S1), although the penA gene did not contain motifs associated with resistance. Gonococci from two of our cases were resistant to multiple antimicrobials. These possessed the gonococcal genetic island, a type IV secretion system known to secrete single-stranded DNA and promote recombination.33 It is therefore possible that acquisition of resistance genes by meningococci could be enhanced in such situations; indeed, urogenital meningococci possessing gonococcal plasmids have been described.34
The prevalence of colonisation with meningococci is likely to be underestimated, as routine methods are optimised to recover gonococci rather than meningococci from sexual health samples. Furthermore, pharyngeal meningococci were not available for comparison. Nevertheless, data in this study show that urogenital/rectal meningococci are diverse and that the majority of these were encapsulated and belonged to hyperinvasive lineages. Urogenital/rectal meningococci are worthy of more attention as sexual transmission may lead to the dispersal of potentially invasive strains.
Key messages
Urogenital/rectal colonisation by Neisseria meningitidis is associated with encapsulated members of hyperinvasive lineages, which contrasts with nasopharyngeal carriage where the prevalence of such meningococci is low.
Although urogenital/rectal carriage with N. meningitidis may be asymptomatic, colonising organisms have the capacity for invasion.
Co-colonisation of potentially invasive meningococci with multidrug-resistant gonococci may promote the transfer of antimicrobial resistance.
Acknowledgments
The authors would like to thank Dr james Bray for help with the genome assembly, Dr Heike Claus for curation of PorB and Matthew Longbone for help with data collection.
References
Footnotes
Handling editor Jackie A Cassell
Contributors OBH, MCJM and JPa designed the study; KC, JPe, DWE, FC and GD collected the isolates and provided sample metadata; OBH and KC did the laboratory work and DNA extractions; OBH did the bioinformatics analyses; OBH, MCJM and JPa analysed the data. OBH prepared the figures and the first draft of the manuscript, which was revised by all the authors.
Funding This work was funded by the Wellcome Trust and the Oxford Martin School (087622/Z/08/2 and H2RXJo00). Additional funding was also provided by the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare-Associated Infections and Antimicrobial Resistance at the University of Oxford in partnership with Public Health England (HPRU-2012-10041) and the NIHR Oxford Biomedical Research Centre. DWE is an NIHR Clinical Lecturer.
Competing interests None declared.
Ethics approval Urogenital N. meningitidis isolates were obtained during background work for a gonorrhoea study15 ‘Using whole genome sequencing to investigate gonorrhoea’ (REC Reference (14/LO/0435) that had received approval from the Brighton and Sussex Research Ethics Committee. Individual consent for the use of anonymised bacterial isolates was not required.
Provenance and peer review Not commissioned; externally peer reviewed.