-
PDF
- Split View
-
Views
-
Cite
Cite
Kelly T. McKee, Timothy M. Shields, Pamela R. Jenkins, Jonathan M. Zenilman, Gregory E. Glass, Application of a Geographic Information System to the Tracking and Control of an Outbreak of Shigellosis, Clinical Infectious Diseases, Volume 31, Issue 3, September 2000, Pages 728–733, https://doi.org/10.1086/314050
Close - Share Icon Share
Abstract
A personal computer—based commercial geographic information system (GIS) was applied to an outbreak of Shigella sonnei infection at Fort Bragg, North Carolina. We used a database consisting of demographic, temporal, and home-address information for all recognized cases of S. sonnei that occurred among health care beneficiaries from 23 May 1997 through 14 August 1997. We imported this database into the GIS, which contained a digitized basemap of the local community. Through simultaneous examination of temporal and spatial distribution of the 59 identified cases of S. sonnei, a focus of infection in a single housing area was identified. Targeted education among residents of the neighborhood in which there was intense transmission was associated with prompt extinction of the epidemic. A GIS offers an efficient and practical way to directly visualize the dynamics of transmission of infectious diseases in the setting of a community outbreak.
Community outbreaks of infectious diarrhea are important public health problems worldwide. Prompt identification of the causative organism; the source, mechanism, and extent of spread; and the characteristics of those affected are critical to timely intervention and institution of control measures.
Infections caused by Shigella species are major causes of both endemic and epidemic diarrhea [1, 2]. The majority of Shigella-related morbidity occurs in developing nations [2]. In the United States, the incidence of shigellosis is disproportionate among Native Americans, institutionalized mentally retarded individuals, and children in day-care centers [2–7].
During the summer of 1997, an outbreak of shigellosis occurred among military health care beneficiaries, including a large proportion of children, at Fort Bragg, North Carolina. The initial investigation found that there was no clear association of disease with any particular child development center or group. The outbreak persisted despite a widespread campaign of handwashing and hygiene awareness.
Coincident with the occurrence of the outbreak, we had begun a demonstration project using a geographic information system (GIS) to map sexually transmitted diseases at Fort Bragg. We speculated that mapping the distribution of identified cases of shigellosis in the population might provide some insight with regard to its source or persistence. In the present study, we describe application of GIS technology to the outbreak, resulting in the prompt identification of a shigellosis “core.” The information that we developed allowed for a targeted environmental assessment, a risk analysis, and a control effort that was temporally associated with extinction of the outbreak.
Patients and Methods
Setting. Fayetteville and nearby Fort Bragg are located in the predominantly rural sandhills region of southeastern North Carolina. Although housing on the military installation is abundant, many active-duty soldiers and their families, as well as other military medical beneficiaries (e.g., retirees), reside within Fayetteville (1997 population, 110,700) and surrounding areas. Primary medical care for military health care beneficiaries is provided through a network of outpatient health clinics associated with Womack Army Medical Center (WAMC) at Fort Bragg. Diagnostic microbiology support is provided by the central clinical laboratory in the Department of Pathology at WAMC.
Case patient definition. Case patients were defined as those individuals who had a stool culture that was positive for Shigella. Individuals who had diarrhea—with or without fever, abdominal pain, nausea, or vomiting—and asymptomatic contact with individuals with known or suspected shigellosis had stool cultures processed for enteric bacterial pathogens by means of standard operating procedures of the Clinical Microbiology Laboratory at WAMC. One additional child with a highly compatible clinical syndrome, who was treated empirically but who did not have culture specimens taken, was considered a case patient for purposes of analysis.
Specimens were collected and were placed into vials containing commercial fecal transport medium (Para-Pak Enteric Plus System; Meridian Diagnostics, Cincinnati), according to the manufacturer's instructions. Enteric pathogen culture media, including MacConkey and Hektoen enteric agars, were inoculated with stool samples and were incubated in the appropriate atmosphere (one that did not contain CO2, in the case of MacConkey and Hektoen) for 18–24 h [8]. MacConkey plates were examined for the presence of non– lactose-fermenting (colorless) colonies, whereas Hektoen plates were examined for the presence of clear or green (Shigella species) colonies or black (Salmonella species) colonies.
Suspicious colonies were subcultured to sheep blood agar and MacConkey agar for another 24 h and were identified by use of the API 20E system (bioMerieux Vitek, Hazelwood, MO). Antimicrobial susceptibilities were determined for organisms that were coded as pathogens by the API 20E system, with use of a MicroScan panel (Dade MicroScan, West Sacramento, CA). Serotyping of Shigella species was performed via agglutination with commercially available antisera (BD Biosciences, Sparks, MD).
Geographic analysis. Mapping of cases of disease was accomplished with MapInfo software (MapInfo, Troy, NY) loaded onto a 260-MHz Pentium-based desktop computer equipped with an inkjet plotter. MapInfo software is a vector-based mapping system with address-matching capability. Commercially available digitized basemaps of Cumberland County, NC, which includes Fayetteville and housing areas on the Fort Bragg installation, were used for mapping purposes. The basemaps included information on street, highway, railroad, and walkway locations; bodies of water; major census-block groups; incorporated and unincorporated locations (as of 1990); 5-digit zip code regions; transportation zones; and regional planning districts. We had previously modified the street basemap file to include a field of building numbers for structures located at Fort Bragg, since many addresses on the installation were provided only as a building number, rather than as a street number and name. Addresses within the study area were mapped by means of addresses (or building numbers) and zip code fields extracted from patient records and encoded within a relational database. Geographic locations of addresses were determined by linear interpolation between street intersections.
Statistical analysis. Space-time analyses of cases of shigellosis were performed by k–nearest neighbor calculations (for which k denotes order of neighbor analysis, i.e., k1 denotes nearest neighbor, k2 denotes next nearest neighbor, etc.) [9], with place of residence as the referent location. The k–nearest neighbor analysis was used under the assumption that cases of shigellosis were locally contagious. Thus, even if the original source of the outbreak could not be identified, a statistically significant result would indicate likely sites where transmission was now established.
Under the statistical null model, it was assumed that cases that were geographically close to one another occurred at random times throughout the outbreak. Rejection of the null hypothesis would indicate that cases that were geographically close to one another also occurred closer together in time than they would have occurred by chance alone. The null model was generated with the use of an approximate randomization of the Mantel product [9] by permutation of the space-time matrix for 500 trials. Distances between cases were estimated as the Euclidian distance between address locations. The geographic locations were estimated as state plane coordinates (North American Datum 1983).
Results
From 23 May through 14 August 1997, 59 cases of shigellosis were identified among health care beneficiaries at Fort Bragg. A total of 58 isolates of Shigella were recovered from stool samples submitted for analysis. Fifty-six isolates were from symptomatic individuals, and 2 were from asymptomatic family members. All isolates were characterized by the laboratory as S. sonnei; in all but 7 cases, reference to a specific serogroup (i.e., group D) was made.
Antimicrobial susceptibility testing revealed that, of the 58 isolates analyzed, 54 were resistant to ampicillin (93%), 55 were susceptible to trimethoprim-sulfamethoxazole (95%), and all 58 were susceptible to ciprofloxacin. Among isolates tested, 100% were susceptible to cefuroxime (55/55), cefotetan (40/40), gentamicin (16/16), and tobramycin (16/16). Twelve (80%) of 15 tested were resistant to cephalothin, and 8 (50%) of 16 displayed intermediate resistance to ticarcillin/clavulanate. All patients were treated successfully with ciprofloxacin (adults only) or trimethoprim-sulfamethoxazole.
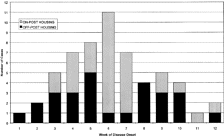
Cases of S. sonnei infection were first identified in late May, with the peak incidence occurring in late June (figure 1). The epidemic apparently began among individuals residing in the local community surrounding the Fort Bragg military installation (off-post residents), because the first cases identified among residents of the installation (on-post residents) began nearly 3 weeks later. The epidemic pattern was bimodal among off-post residents (weeks 3–5 and 8–10), whereas there was a strong single peak among cases occurring on post (weeks 6 and 7). The ages of those individuals who were identified as being infected ranged from 1 to 43 years (mean, 13.13 years). Forty-six percent of infected persons were male. Cases tended to cluster in households; 38 (64%) of 59 cases occurred at residences from which more than 1 positive culture specimen was obtained.
Fifty-three of the 59 individuals infected with S. sonnei (90%) resided in Cumberland County, North Carolina, at the time of diagnosis; of these, 52 could be geocoded. Approximately half of the identified cases (30 of 59) occurred among beneficiaries living on the Fort Bragg installation. During peak epidemic weeks, most cases occurred among on-post residents (figure 1).
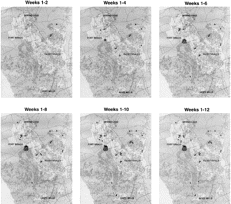
Analysis of the geographic distribution and dates of onset of the cases indicated significant space-time clustering of cases (PJ[k] = .002 for neighboring cases 1, 2, and 3, where PJ[k] denotes the probability that the total number of cases is significantly clustered; Simes correction, P = .02) (figure 2). Thus, for any case, the nearest 3 cases were more likely to be associated in space and time than would be expected by chance. Occurrence of space-time clustering was taken as an indication of local transmission of infection, even though a single source could not be identified.
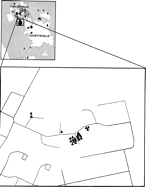
We were able to identify focus of infection in a single housing area on the Fort Bragg installation (figure 2). On closer examination of this area, it became apparent that most cases in this geographic cluster were occurring among residents of a “microneighborhood,” which was defined by several streets within this subdivision (figure 3). Armed with this information, a small team of disease-intervention specialists was dispatched to this area. Interviews with case families and neighbors revealed that small communal wading pools had been set out in certain yards and had been frequented by young children from most affected families. With cessation of this activity and introduction of home-based information campaigns and education, further spread within this area was stopped.
Discussion
GISs are being applied, with increasing frequency, to public health practice, planning, and research efforts [10]. GIS technology provides for the rapid integration and analysis of data from multiple sources and types, with visual display of information in space and time. Using a relatively inexpensive, off-the-shelf GIS system operating on a desktop computer system, we were able to monitor the occurrence and evolution of an outbreak of diarrheal disease in our local community. The information we obtained through application of the GIS enhanced our understanding of the dynamics of the epidemic and helped us identify an intervention target.
This epidemic of S. sonnei infection occurred in 2 phases. Cases initially were recognized among health care beneficiaries living off post and were observed in several different locations. The first cases among persons living on post were scattered as well, but once the outbreak became established in a single neighborhood, a second, rapidly progressive local epidemic occurred.
In the United States, person-to-person contact is believed to be the principle means by which shigellosis is spread [2, 11, 12], although outbreaks have also been attributed to contaminated food and water [1, 13–15] and to exposure to recreational water sources [1, 16]. Most domestically acquired S. sonnei infections occur in children <10 years old [12, 17]. In families, the index case is usually a child [17, 18]. The index case of the outbreak in the “core” neighborhood identified by GIS was a 43-year-old man. The infection spread to his family members and, subsequently, to nearby residences. We speculate that contamination of water in neighborhood wading pools was a major contributor to interfamilial transmission in this “core” neighborhood; spread of the infection within families and among individuals outside this neighborhood most likely occurred via more conventionally recognized interpersonal spread.
A common source for this outbreak of S. sonnei was never proven microbiologically; no attempts were made to examine plasmid profiles or to perform other molecular analyses to document uniformity among isolates [19–22]. However, the occurrence of large numbers of cases of dysentery in which strains of S. sonnei with very similar antibiotic susceptibility patterns were recovered, together with our finding of significant space-time clustering of cases, strongly suggests the epidemic spread of a single strain. Circumstantial evidence suggests that the outbreak among military health care beneficiaries occurred as part of a larger, countywide epidemic. The second wave of off-post cases, which occurred after week 7 (figure 1), was found in areas distinct from those where the initially recognized group of cases was found during weeks 1–5 (figure 2). The cases that occurred on post after week 7 were sporadic and spatially scattered, suggesting absence of an ongoing, active transmission focus at Fort Bragg.
As the outbreak unfolded, we became increasingly frustrated by the lack of a discernable pattern of spread. Affected infants and young children attended several different child development facilities (both on and off post); a variety of different water sources were used by affected families; and no common gathering, foodstuff, or eating establishment could be identified. Our GIS analysis enabled us to demonstrate significant space-time clustering of cases, suggesting the occurrence of local transmission; in addition, a temporal and spatial perspective of the outbreak was provided, enabling us to paint a comprehensive picture of the evolution and distribution of the epidemic. A localized area of high incidence of disease was readily identifiable, and a target for intervention activities—which, up to that point, had been generic and unfocused—was provided. We cannot be certain that the interventions that were used affected the course of the outbreak. However, shortly after the localized area of disease activity was noted and the use of community wading pools was discouraged, the outbreak tapered off.
The utility of the GIS in the tracking and control of this outbreak of S. sonnei was heightened by the occurrence of a relatively large number of bacteriologically confirmed cases during a short interval. However, the direct application of GIS technology to the control of disease in settings in which most cases in the community may be microbiologically undetected and of limited number (e.g., as with endemic shigellosis) might prove more problematic. One possible approach to this situation involves integration of a provider reporting/notification network with GIS-based surveillance; such a system could allow for rapid initiation of case-finding and intervention on the basis of syndromic re-porting.
GISs are finding increasingly widespread application in the prediction, management, and control of infectious diseases and other public health problems [23–25]. Integration of GIS technology into the routine monitoring of health events offers a rapid and quantitative method for identification of unusual disease patterns and also provides a vehicle for disease investigation as well as a tool for measuring the success of interventions. For communities with health care systems supported by automated ambulatory or inpatient disease recording, creation of an interface between a GIS and the disease data system can be relatively simple and inexpensive.
GIS technology may realize its greatest potential in the setting of diseases with high epidemic potential occurring under natural or artificial conditions (e.g., purposeful spread of a biological weapon). Integration of a syndrome-based reporting system with community GIS monitoring provides the opportunity to rapidly recognize and localize the occurrence of disease cases or unusual disease patterns. When a GIS is coupled with meteorological, transportation, topographical, or other data systems, patterns of spread can be readily appreciated, and predictive models can be generated to facilitate intervention. Thus, for surveillance programs faced with the prospect of the introduction of exotic disease or bioterrorism, GIS technology could readily serve as the cornerstone for the recognition of an event and prompt intervention.
Acknowledgment
We wish to acknowledge the support and assistance provided by the staffs of the Preventive Medicine Service and Clinical Microbiology Laboratories at WAMC.
References
Figures and Tables
Epidemic curve of the outbreak of shigellosis, which occurred from 23 May through 14 August 1997, at Fort Bragg, North Carolina. Light bars, Cases occurring among health care beneficiaries residing on post. Dark bars, Cases occurring among beneficiaries residing off post.
GIS maps of residential address locations where cases of shigellosis occurred among Womack Army Medical Center health care beneficiaries in Cumberland County, North Carolina, from 23 May through 14 August 1997. The successive figures display the cumulative distribution of cases at 2-week intervals during the outbreak.
GIS display of a neighborhood containing a large cluster of cases of shigellosis. Note the predominance of cases grouped among families living in close proximity to each other.
Presented in part: 37th Annual Meeting of the Infectious Diseases Society of America, Philadelphia, 18–21 November 1999 (abstract no. 585).
Financial support: US Army Center for Health Promotion and Preventive Medicine (Aberdeen Proving Ground, MD) and the Department of Defense Global Emerging Infections Surveillance System (Washington, D.C.).
The views and opinions expressed herein are those of the authors and do not necessarily reflect the official policy position of the US Army, the Department of Defense, or the Henry M. Jackson Foundation for the Advancement of Military Medicine.
None of the authors of this study have commercial or other associations that might represent a conflict of interest.
Author notes
Present affiliations: US Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD (K.T.M.); The Care Clinic, Fayetteville, NC (P.R.J.).





Comments